

The R CMD BATCH command has a lot of options you could specify, mostly manipulating how your R session is configured. > t.test(mpg ~ am, data=mtcars, var.equal=TRUE)Īlternative hypothesis: true difference in means is not equal to 0 'help.start()' for an HTML browser interface to help. Type 'demo()' for some demos, 'help()' for on-line help, or
#R studio commands regression how to#
'citation()' on how to cite R or R packages in publications. Type 'contributors()' for more information and R is a collaborative project with many contributors. Natural language support but running in an English locale Type 'license()' or 'licence()' for distribution details. You are welcome to redistribute it under certain conditions.
#R studio commands regression software#
R is free software and comes with ABSOLUTELY NO WARRANTY. In the Files pane you can find the output file and open it:Ĭopyright (C) 2020 The R Foundation for Statistical Computing Then issue this command in the Console: tools::Rcmd("BATCH -no-save testscript.R")
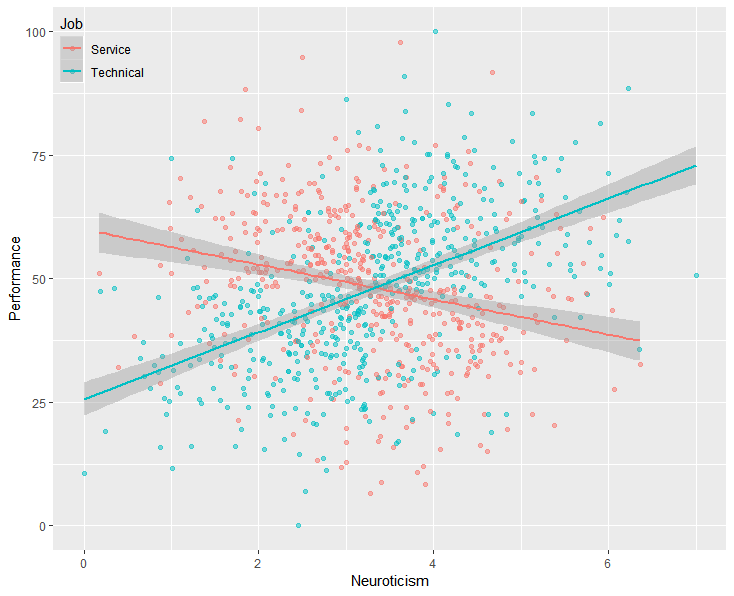
T.test(mpg ~ am, data=mtcars, var.equal=TRUE) Then you process that file.įor example, save a file (our example is called testscript.R) with the following commands in your working directory: # testscript.R, used as an example If you want to save a large quantity of output that includes the commands that produced it, you really want BATCH output (Stata-style output). However, this quickly gets complicated, and can be difficult to interpret. It is also possible to sink() error messages by adding the argument type = "message". Which saves the following text in a file called “outputfile.txt”:Īlternative hypothesis: true mean is not equal to 0 Use one sink() command with a filename to begin saving output, and another empty sink() command to stop. If you are running many similar commands with similar output, using this approach to create a single file quickly becomes difficult to read. This gives you clean output (SAS-style), and might be suitable for producing a simple report. This redirects your output to a file, while commands and error messages continue to go to the console. One option is to use the sink() function. Refer to the earlier discussion of the working directory for help finding and setting the working directory. When we use batch processing, we need to ensure our script (“testscript.R”) is saved in our working directory so that R can find it we will then find the output file (“testscript.Rout”) in our working directory as well. When we use sink(), we will find the output file (“outputfile.txt”) in our working directory. Here we will consider two basic methods that you might use when working within RStudio.Įach of these methods will save and read files from our working directory. There are numerous methods of capturing printed output automatically. Some cases call for saving all of our output, such as creating log files. In most cases, you should be intentional about how you save output, such as saving datasets as RDS files and regression results as formatted tables with the stargazer package. However, the Console only buffers a limited amount of output (1000 lines by default), making it difficult to work with large quantities of output. When working in the RStudio, R echoes commands, prints output, and returns error messages all in one place: the Console. 10.1 What packages are already installed?.


 0 kommentar(er)
0 kommentar(er)
